pStab
Prediction of Stable Mutants, Unfolding Curves, Stability Maps and Protein Electrostatic Frustration
Hot-Spots and Frustration
∆Eelec Distribution
Thermal Unfolding Curves
Local Stability Profile
Input parameters
pH = 7 T = 298 K Ionic strength = 0.1 M No. of allowed mutations = 3
Charged/Polar residues to be excluded = None
NOTE: Hover mouse over data-point to see value and select area to zoom.
?
NOTE: Use your mouse to drag, rotate, and zoom in and out of the structure.
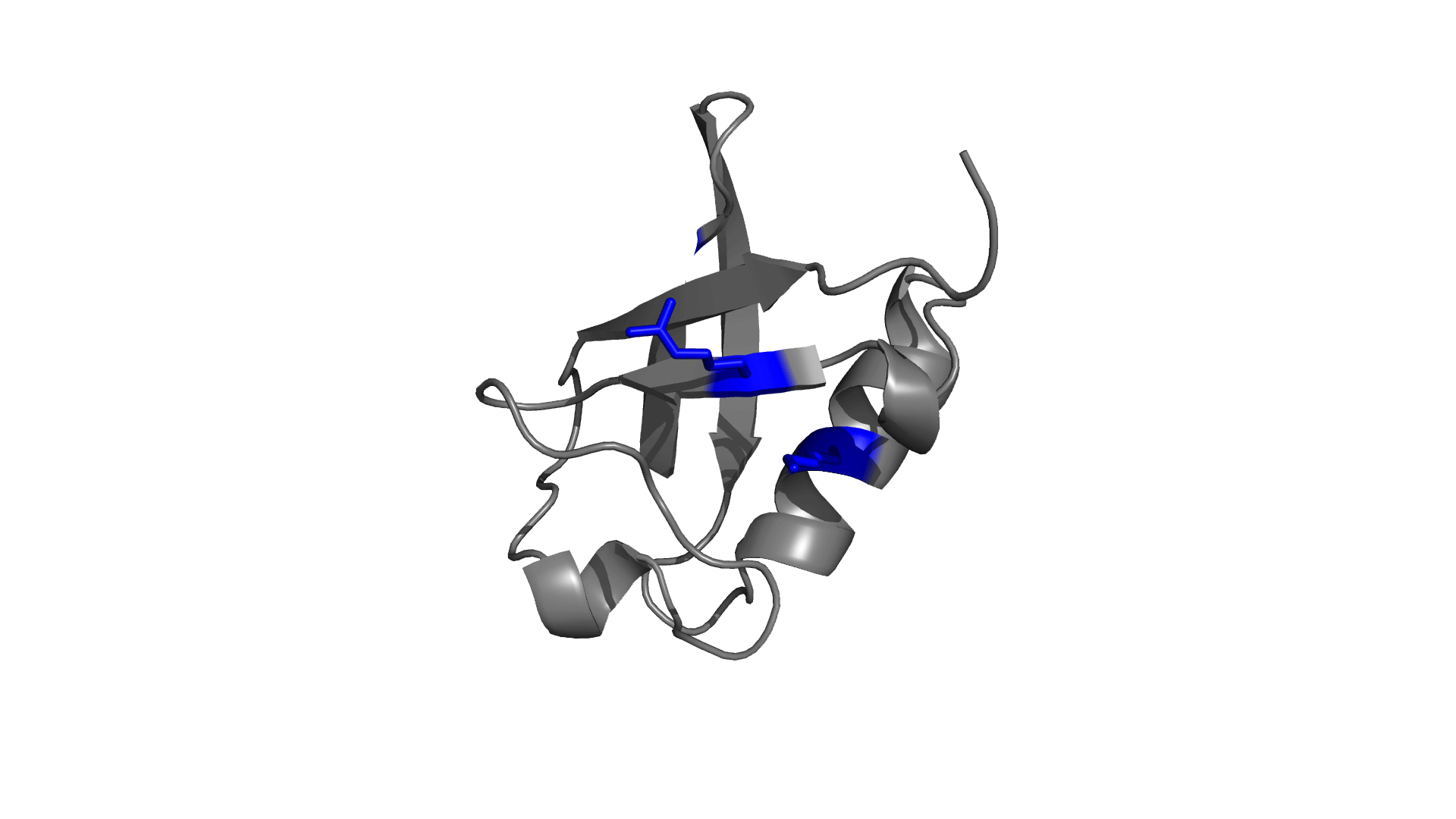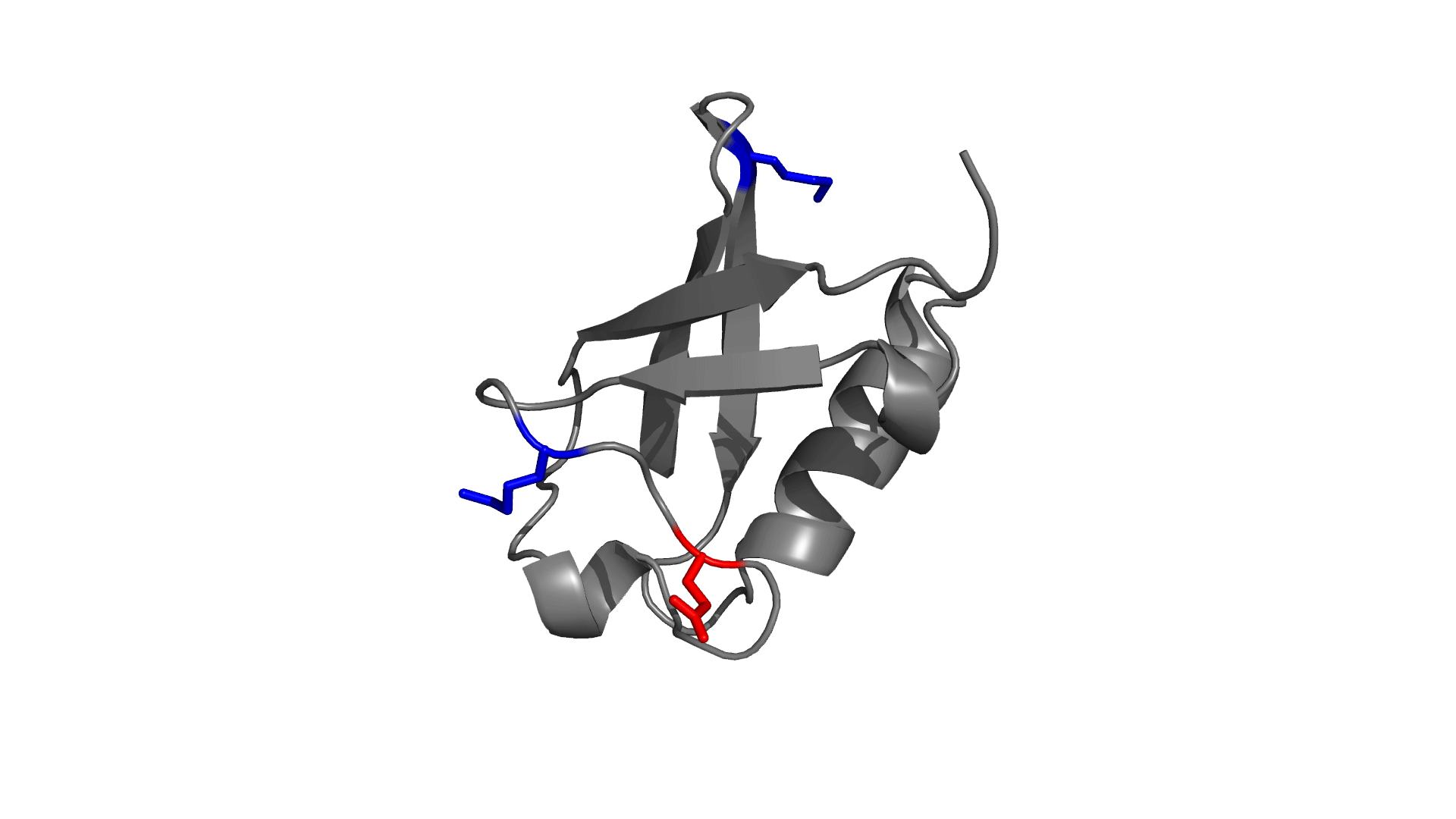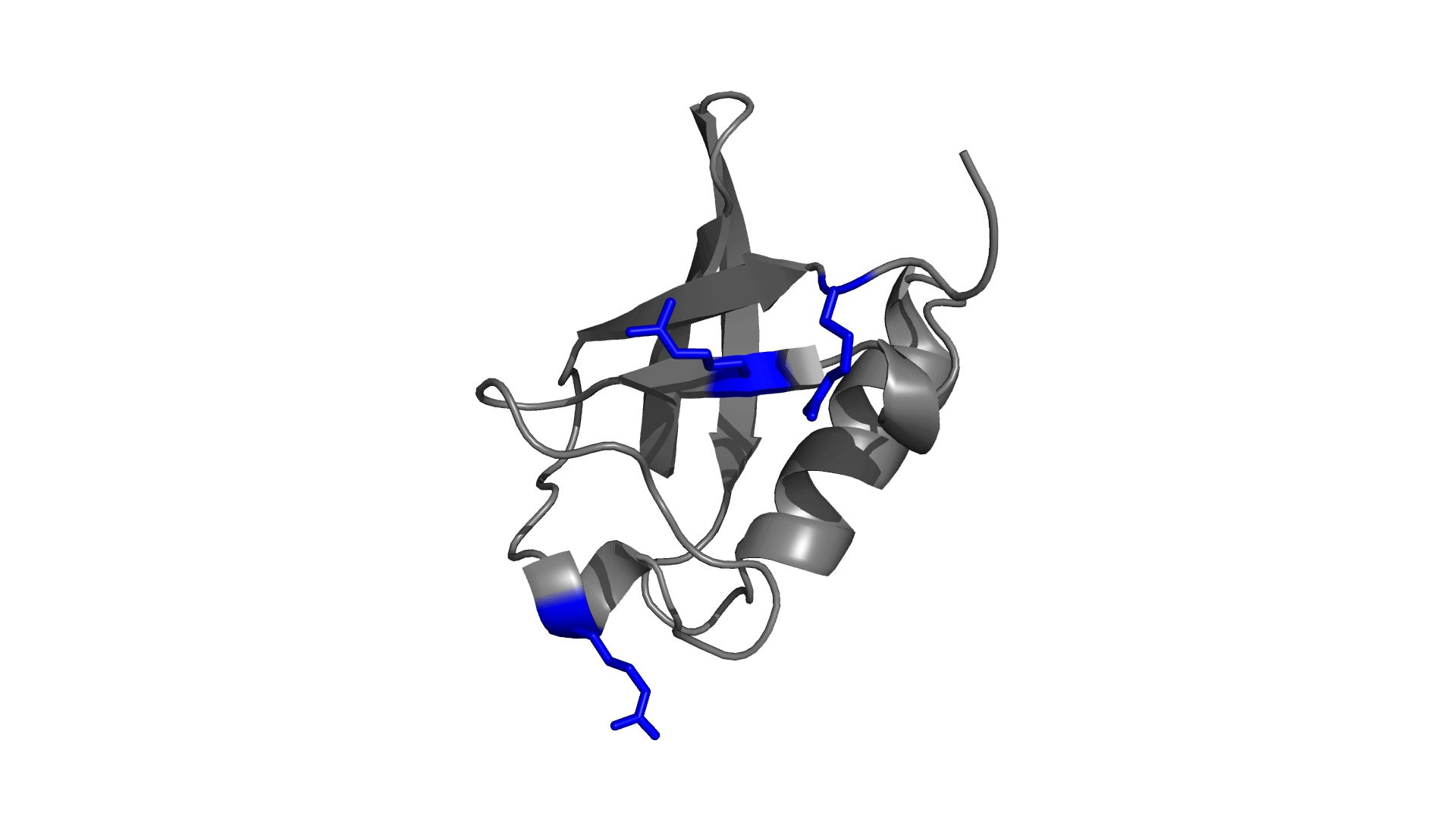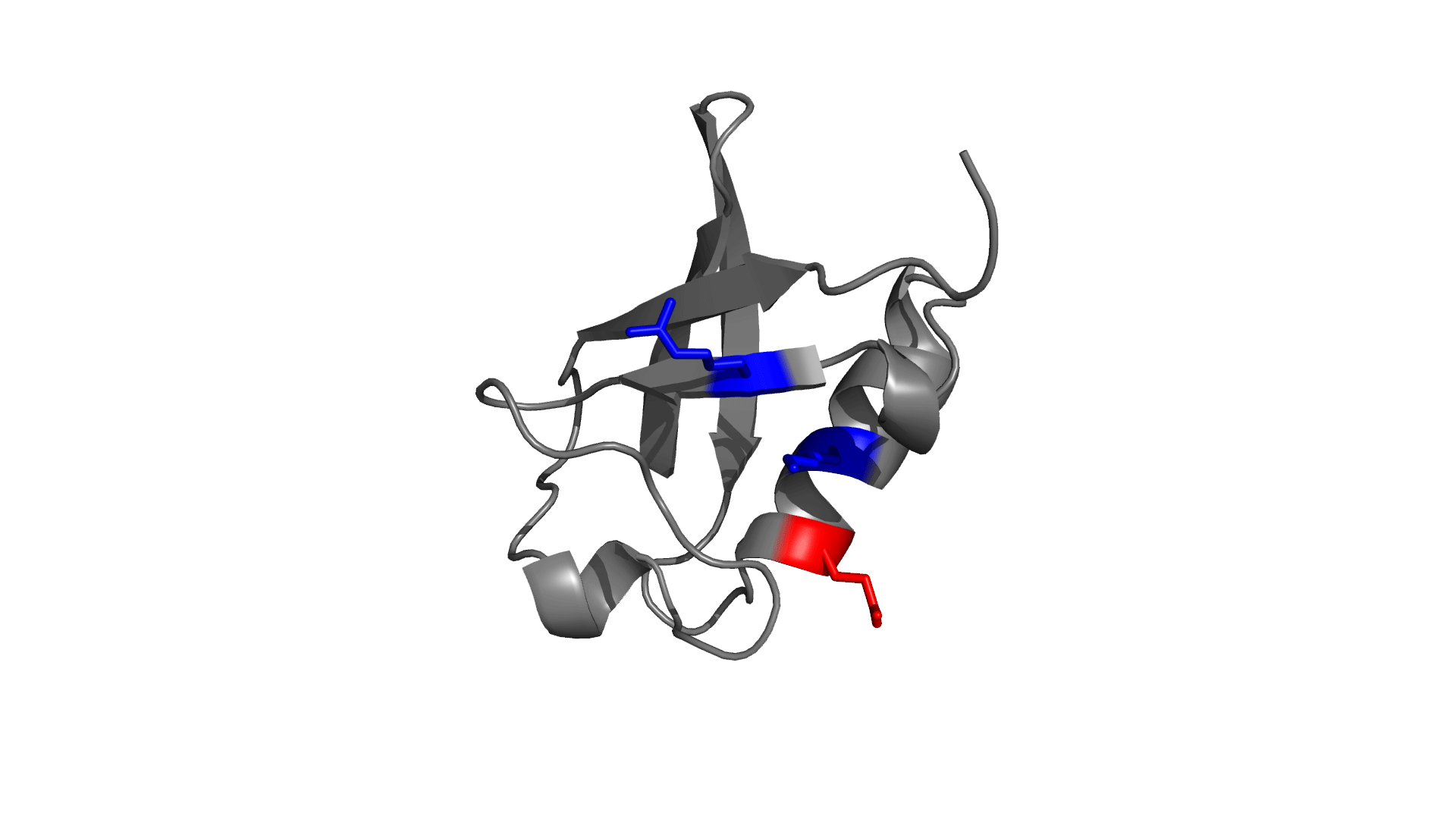
Positively charged / Negatively charged
Favorable / Unfavorable
Download the electrostatic frustration map of wild type protein.
D O W N L O A D
Input parameters
pH = 7 T = 298 K Ionic strength = 0.1 M No. of allowed mutations = 3
Charged/Polar residues to be excluded = None
?
NOTE: Click on legend to show/hide data-series.
| No. of Substitutions | No. of Mutants Generated | No. of Stable Mutants |
|---|---|---|
| Single Mutants | 44 | 12 |
| Double Mutants | 924 | 141 |
| Triple Mutants | 12320 | 1030 |
| All Mutants | 13288 | 1183 |
Download the list of top 5000 stable mutants.
D O W N L O A D
Input parameters
Exp. Tm of WT = 333 K pH = 7 Temperature = 298 K Ionic strength = 0.1 M
Entropic cost: Secondary structure based entropic cost
Method: Generate mutant(s) by virtual mutagenesis
NOTE: Click on legend to show/hide data-series.
?
| Mutant Index | Protein Variant | Tm (K) | ΔTm (K) |
|---|---|---|---|
| - | Wild-Type | 333.6 | - |
| 1 | E18Q/E24K/R42Q | 341.5 | 7.9 |
Download the unfolding curves of wild type and mutant proteins.
D O W N L O A D
Input parameters
Exp. Tm of WT = 333 K pH = 7 Temperature = 298 K Ionic strength = 0.1 M
Entropic cost: Secondary structure based entropic cost
Method: Generate mutant(s) by virtual mutagenesis
NOTE: Click on legend to show/hide data-series.
?
NOTE: Click on Zoom In to visualize trends.
?
Download the local stability profiles of wild type and mutant proteins.
D O W N L O A D
Protein Biophysics Lab

2017, Maintained by Protein Biophysics Lab, IIT Madras, Chennai-36, India
Last Updated: September 13, 2017
