 Protein Biophysics Lab
Protein Biophysics Lab 
pStab
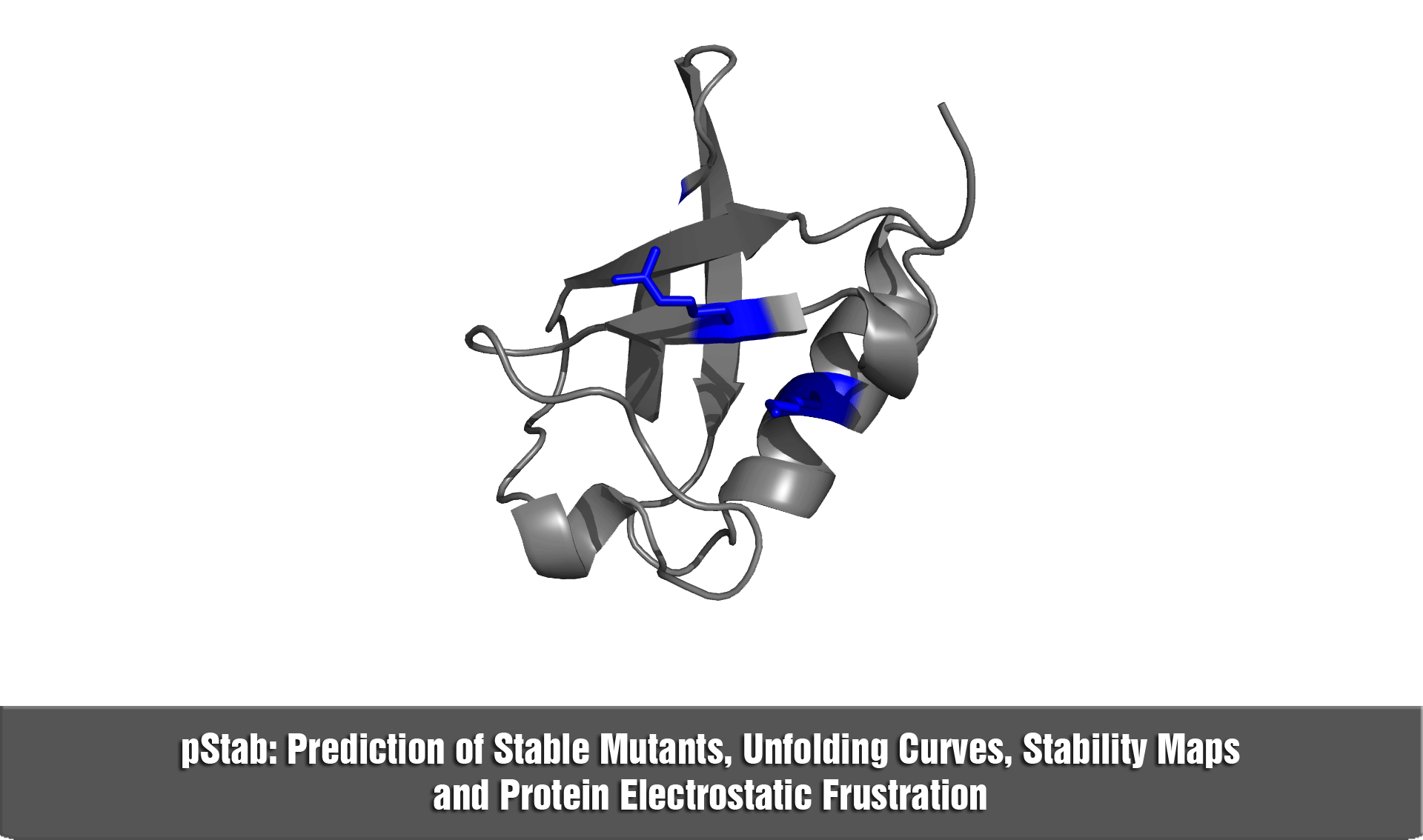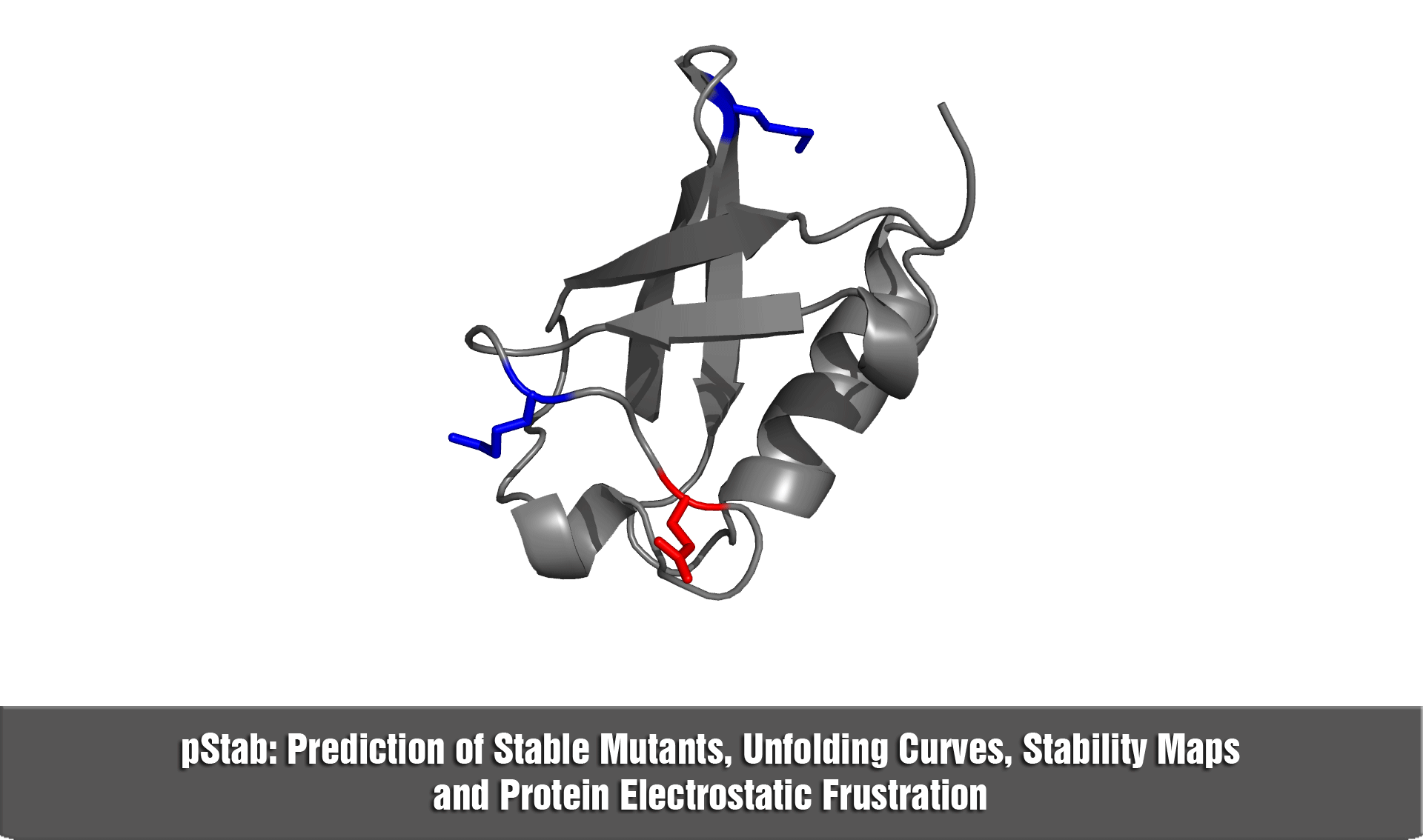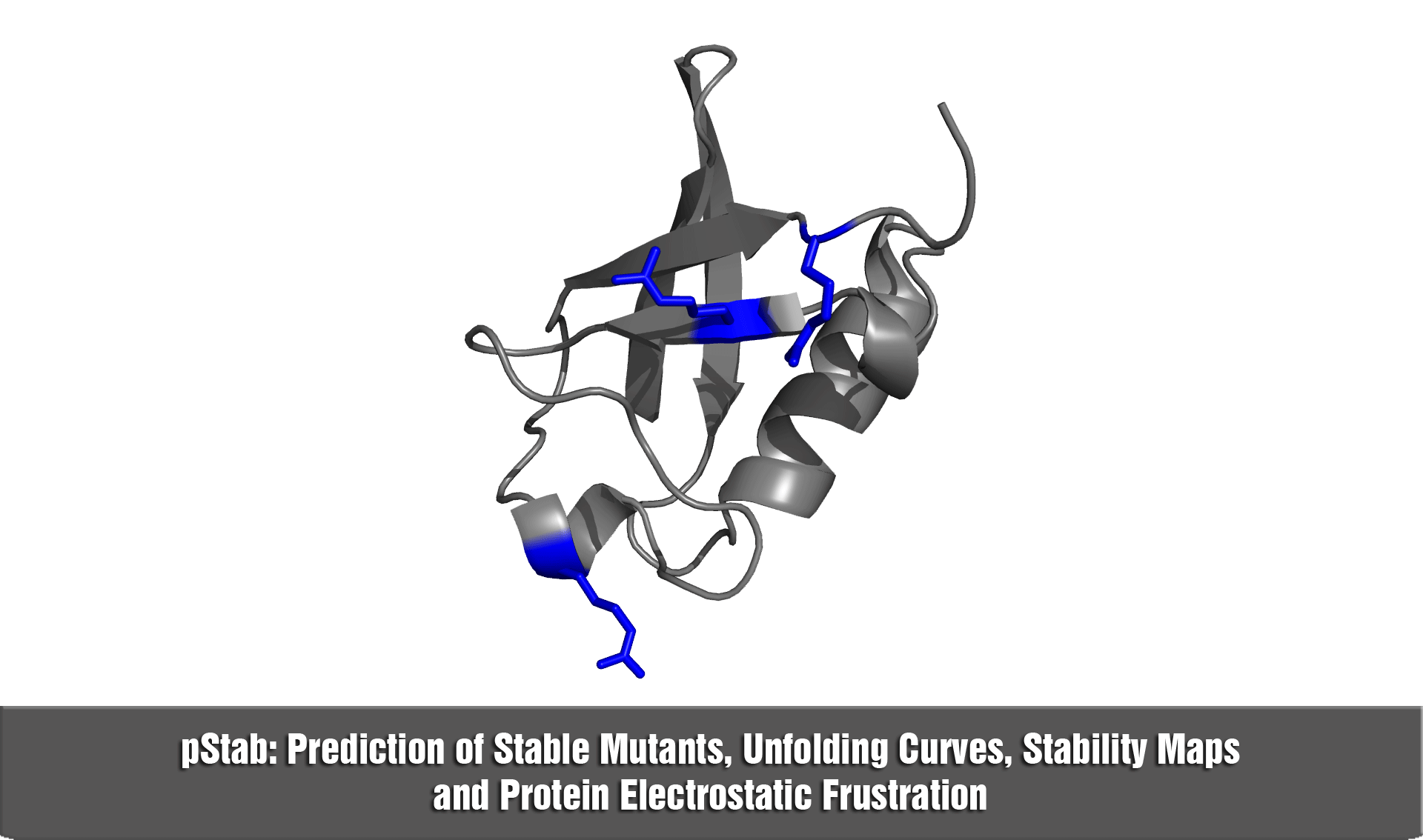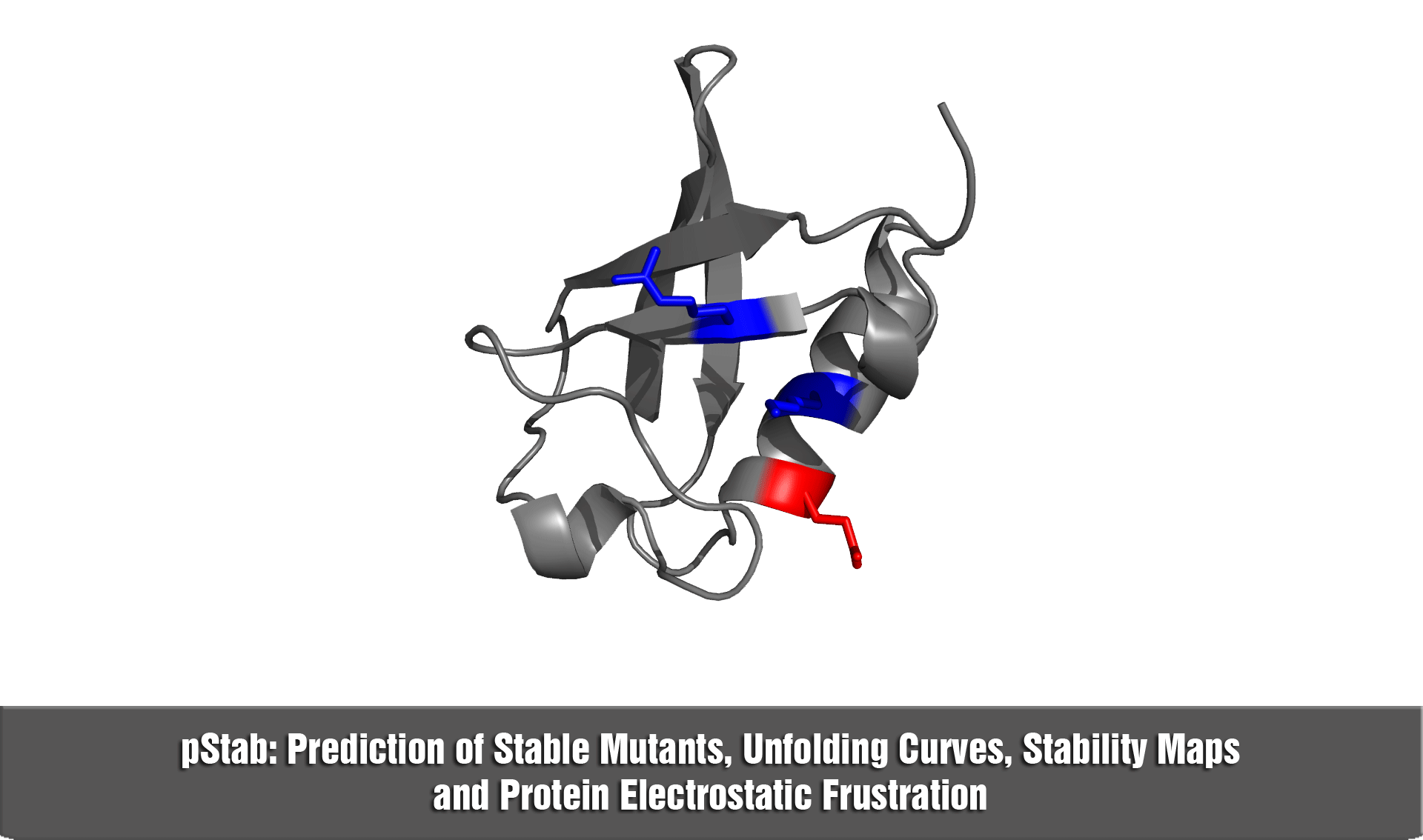
pStab web server provides a robust and fast strategy to engineer protein stabilities through mutations involving charged residues.
We expect the method to be of importance
a) as a first and rapid step to screen for protein mutants with specific stability in the biotechnology industry
b) in the construction of stability maps at the residue level (i.e., hot spots)
c) as a robust tool to probe for mutations that enhance the stability of protein-based drugs
d) to explore the role of electrostatic frustration in proteins.
pPerturb
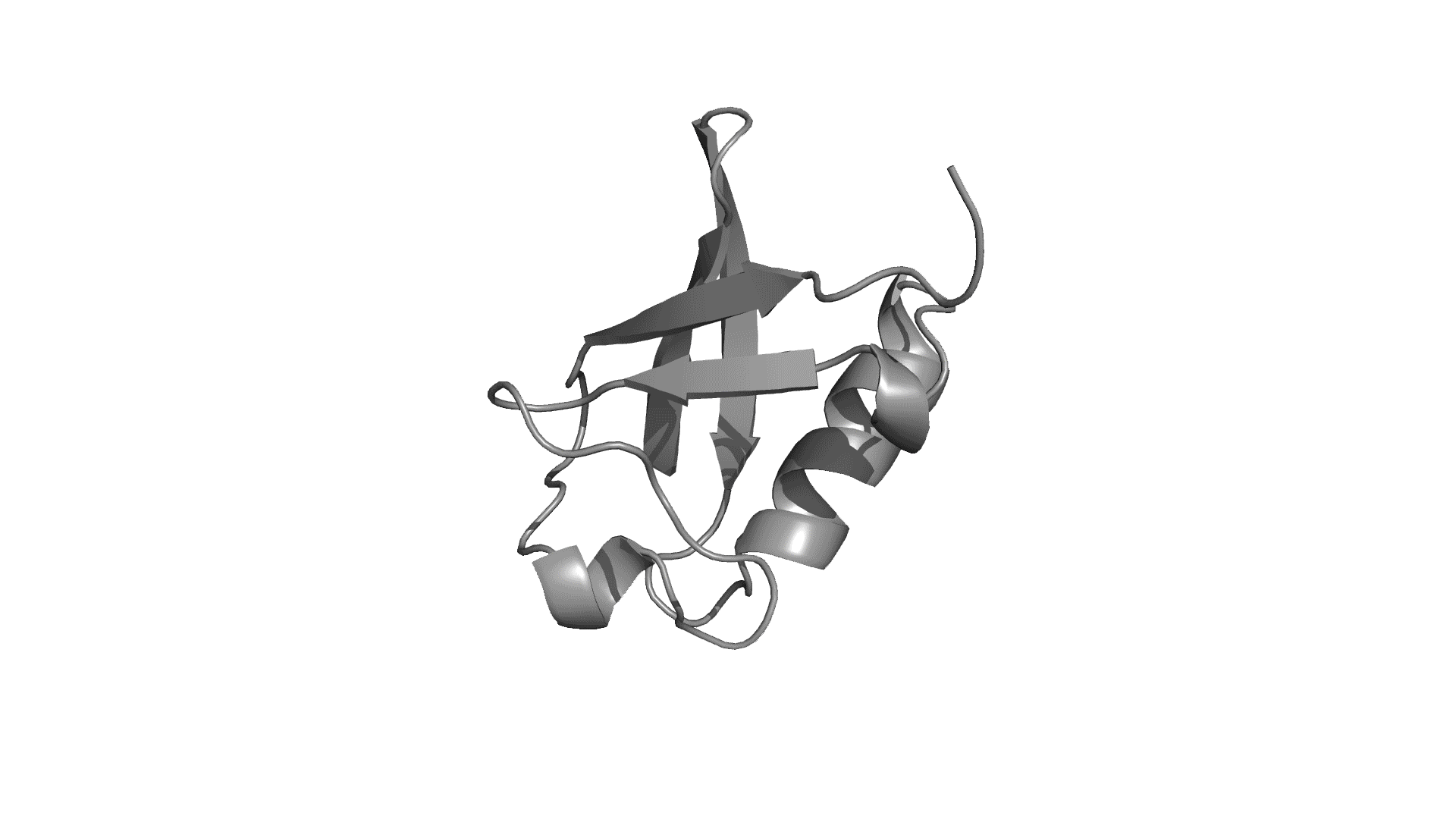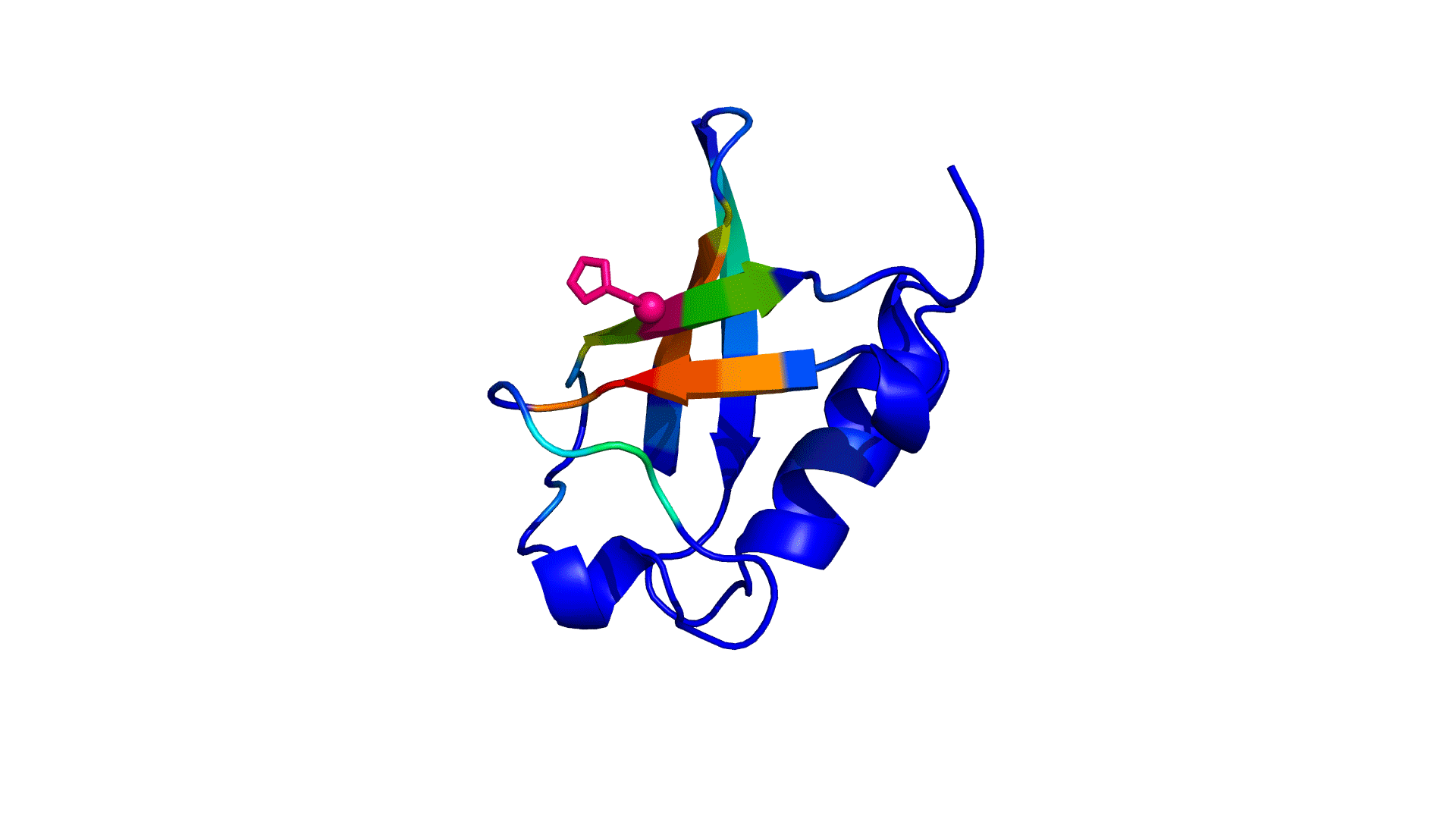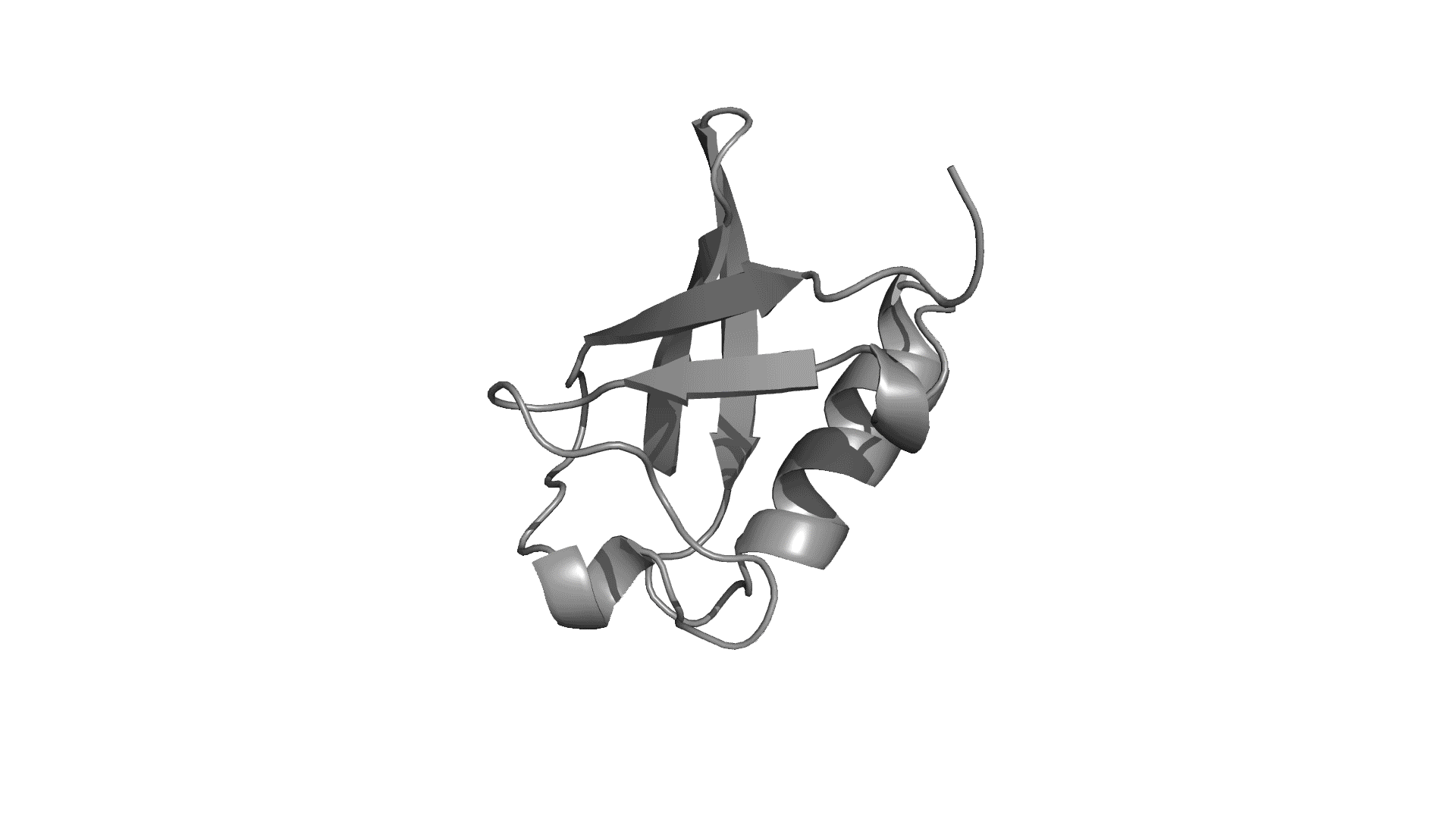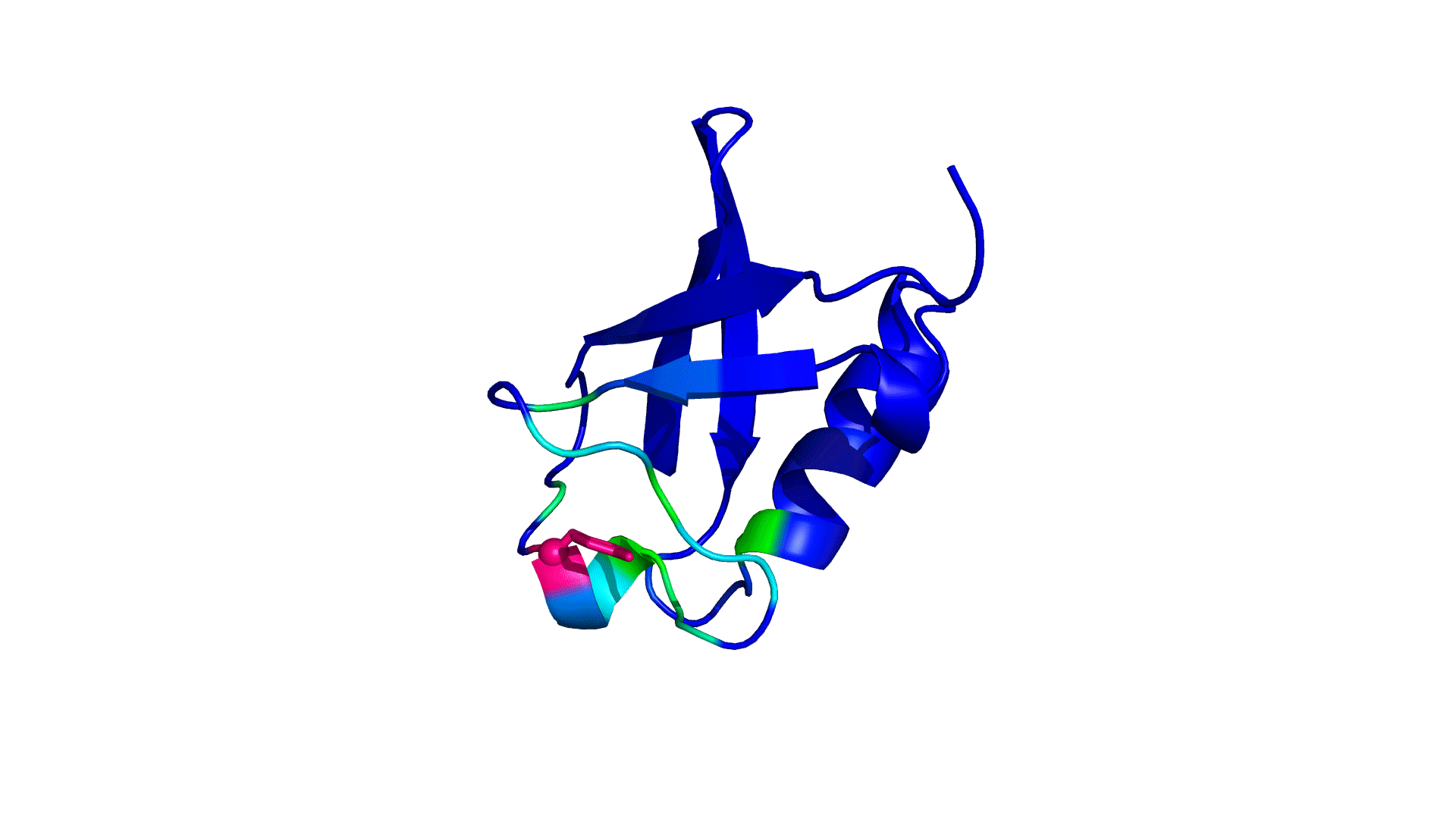
pPerturb web server provides a rapid and experimentally consistent measure of the extent to which a particular residue is coupled to its neighbors.
We expect the method to be of importance
a) to explore the strength of the interaction network around a particular residue
b) in quantifying the partitioning of destabilization energetics around the mutation neighborhood
c) as a simple theoretical framework for modeling allosteric effects
d) to explore the effects of thermodynamics upon side-chain truncating mutations.
2023, Maintained by Protein Biophysics Lab, IIT Madras, Chennai-36, India