
pFolMech
Protein Biophysics Lab

Predicting Folding Free Energy Profiles, Conformational Landscapes, and Folding Mechanisms
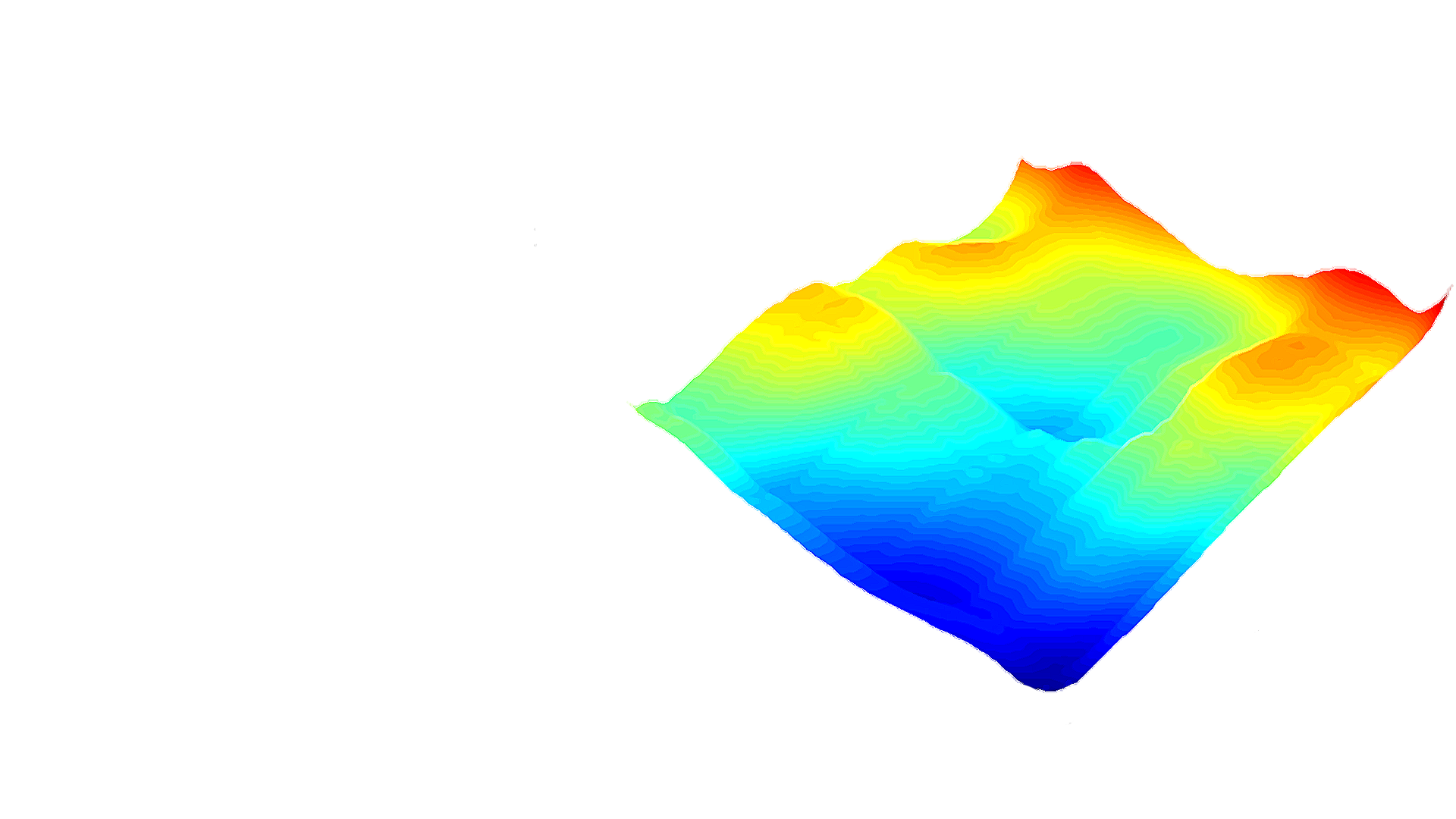
General Instructions
- The protein length should be between 50 – 500 amino acids (Visit help page for more details).
-
The uploaded PDB file
- Should not contain any hetero atom(s) or non-standard amino acid(s).
- Should contain the complete description of heavy atoms (To model missing atoms, visit here).
- Should contain one structural state (i.e. one model entry) in case of NMR structures.
-
Large proteins (>100 amino acids) are approximated using a block definition of block size = ceil(nres/100), where nres corresponds to the number of residues in the protein.
- For example, a protein of length 452 will be split into multiple blocks whose most probable block size is 5.
- Maximum block size of 5 is used.
- The effects of disulphide bridges are not accounted for by the web server.
2022, Maintained by Protein Biophysics Lab, IIT Madras, Chennai-36, India
Last Updated: February 8, 2022